Code
library(tidyverse)This document provides a detailed overview of the fundamental data structures in R. Understanding these structures is essential for effective programming and data analysis in R. Each structure has its own strengths and is suited for different types of tasks.
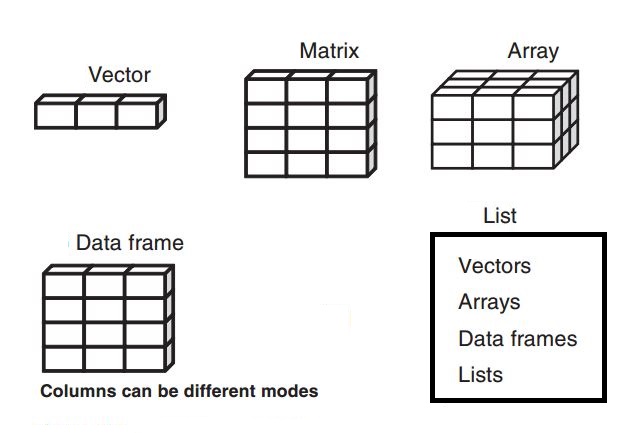
A vector is a one-dimensional, ordered collection of elements. A key characteristic of vectors is that all elements must be of the same data type (homogeneous). They are the simplest and most common data structure in R.
Vectors are typically created with the c() (combine) function.
[1] 10.5 2.3 15.0 4.8[1] "apple" "banana" "cherry"[1] TRUE FALSE TRUE TRUEThe seq() function generates a sequence of numbers.
The rep() function repeats a value a specified number of times.
The sample() function takes a random sample from a set of elements. replace = FALSE means each element can only be chosen once.
With replace = TRUE, elements can be chosen multiple times.
runif() generates random numbers from a uniform distribution.
rnorm() generates random numbers from a normal distribution.
The unique() function removes duplicate elements from a vector.
If you try to mix types, R will coerce the elements to the most flexible type in a specific hierarchy: logical -> integer -> numeric -> character. For example, mixing numbers and characters will result in a character vector because character is the most flexible type.
Mathematical and logical operations on vectors are performed element-wise. This is a powerful feature called vectorization, which is much faster than writing a loop.
You can select elements from a vector using their index inside square brackets []. R uses 1-based indexing.
[1] "banana"[1] 10.5 15.0[1] 10.5 15.0[1] 10.5 15.0 4.8You can combine vectors by using the c() function.
Negative indexing removes elements at the specified positions.
Remove the first element:
Remove the last element:
Remove elements based on a vector of indices:
sort() arranges vector elements in ascending or descending order.
length() returns the number of elements in a vector.
Mathematical functions can be applied to entire vectors.
Select the first element:
Select the last element:
Select a range of elements:
setdiff(x, y) finds elements that are in vector x but not in vector y.
as.* functions are used to coerce vectors from one type to another.
A data frame is a two-dimensional, heterogeneous data structure, similar to a spreadsheet or a SQL table. It is the most important data structure for data analysis in R.
Use the data.frame() function or, preferably, the tibble() function from the tidyverse. Tibbles are a modern take on data frames that are more user-friendly: they don’t change variable names or types, and they have a much nicer print method.
You can subset data frames in several ways:
$ or [[ ]] to select a single column by name. This returns the column as a vector.[row, column] to select specific rows and columns. The result is another data frame (or a vector if you select a single column).[1] "Alice" "Bob" "Charlie" "David" # A tibble: 2 × 2
name score
<chr> <dbl>
1 Alice 95
2 Bob 82For more complex subsetting, it is highly recommended to use the dplyr verbs filter() and select() (covered in a separate guide).
Converting a data frame to a matrix will coerce all elements to the most flexible data type (usually character).
You can extract a single column as a vector using $ or [[ ]] notation.
A matrix is a two-dimensional, homogeneous data structure. All elements must be of the same type. It is most useful for mathematical and statistical operations, such as linear algebra.
Use the matrix() function.
Subsetting is similar to data frames, using [row, column] notation.
Matrices support matrix algebra operations, such as transposition (t()) and matrix multiplication (%*%).
A list is a one-dimensional, heterogeneous data structure. Unlike vectors, lists can contain elements of different types and even different structures, including other lists, vectors, or data frames. This makes them very flexible, like a container for other objects.
Use the list() function. It’s good practice to name the list elements.
$name
[1] "Linear Model"
$coefficients
intercept slope
2.5 0.8
$r_squared
[1] 0.85
$data
mpg cyl disp hp drat wt qsec vs am gear carb
Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4
Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4
Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1
Hornet 4 Drive 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1
Hornet Sportabout 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2
Valiant 18.1 6 225 105 2.76 3.460 20.22 1 0 3 1Subsetting lists requires understanding the difference between [ and [[:
[[index]] or [[name]] extracts a single element from the list. The result will have the type of that element.[index] or [name] extracts a sub-list. The result is always another list, even if you only select one element.intercept slope
2.5 0.8 $coefficients
intercept slope
2.5 0.8 [1] 0.85An array is a multi-dimensional, homogeneous data structure. It can have two or more dimensions. A 2D array is essentially a matrix. Arrays are useful for storing data that has more than two dimensions, such as image data (height, width, color channels) or time series data for multiple locations.
Use the array() function, specifying the data and the dimensions.
Elements are accessed using [row, column, dimension] notation.
Understanding the structure of your data is a critical first step in any analysis. R provides several useful functions for this.
str(): The most useful function. Provides a compact, human-readable summary of any R object.class(): Returns the high-level class of an object.typeof(): Returns the internal storage type.length(): Returns the number of elements in a vector or list.dim(): Returns the dimensions (e.g., rows and columns) of a data frame, matrix, or array.names() or colnames(): Returns the names of elements or columns.tibble [4 × 4] (S3: tbl_df/tbl/data.frame)
$ id : int [1:4] 1 2 3 4
$ name : chr [1:4] "Alice" "Bob" "Charlie" "David"
$ score : num [1:4] 95 82 78 91
$ is_student: logi [1:4] TRUE FALSE TRUE FALSEList of 4
$ name : chr "Linear Model"
$ coefficients: Named num [1:2] 2.5 0.8
..- attr(*, "names")= chr [1:2] "intercept" "slope"
$ r_squared : num 0.85
$ data :'data.frame': 6 obs. of 11 variables:
..$ mpg : num [1:6] 21 21 22.8 21.4 18.7 18.1
..$ cyl : num [1:6] 6 6 4 6 8 6
..$ disp: num [1:6] 160 160 108 258 360 225
..$ hp : num [1:6] 110 110 93 110 175 105
..$ drat: num [1:6] 3.9 3.9 3.85 3.08 3.15 2.76
..$ wt : num [1:6] 2.62 2.88 2.32 3.21 3.44 ...
..$ qsec: num [1:6] 16.5 17 18.6 19.4 17 ...
..$ vs : num [1:6] 0 0 1 1 0 1
..$ am : num [1:6] 1 1 1 0 0 0
..$ gear: num [1:6] 4 4 4 3 3 3
..$ carb: num [1:6] 4 4 1 1 2 1| Use a… | When you have… | Example Use Case |
|---|---|---|
| Vector | A 1D sequence of the same data type. | A single column of data, like ages or names. |
| Data Frame | A 2D table with columns of different data types. | Your typical dataset for analysis. |
| Matrix | A 2D grid of the same data type. | A correlation matrix, an image grayscale matrix. |
| List | A collection of different types/structures. | To return multiple, varied objects from a function. |
| Array | A multi-dimensional grid of the same data type. | 3D medical imaging data, climate data over time. |
---
title: "Data Structures in R"
execute:
warning: false
error: false
format:
html:
toc: true
toc-location: right
code-fold: show
code-tools: true
number-sections: false
code-block-bg: true
code-block-border-left: "#31BAE9"
---
This document provides a detailed overview of the fundamental data structures in R. Understanding these structures is essential for effective programming and data analysis in R. Each structure has its own strengths and is suited for different types of tasks.
{width="600"}
```{r}
library(tidyverse)
```
# 1. Vectors
A vector is a one-dimensional, ordered collection of elements. A key characteristic of vectors is that all elements must be of the same data type (**homogeneous**). They are the simplest and most common data structure in R.
### Creating Vectors
Vectors are typically created with the `c()` (combine) function.
```{r}
# A numeric vector
numeric_vector <- c(10.5, 2.3, 15.0, 4.8)
numeric_vector
# A character vector
character_vector <- c("apple", "banana", "cherry")
character_vector
# A logical vector
logical_vector <- c(TRUE, FALSE, TRUE, TRUE)
logical_vector
```
#### Sequence Vector
The `seq()` function generates a sequence of numbers.
```{r}
seq(from = 2, to = 14, by = 2)
```
#### Replicated Vector
The `rep()` function repeats a value a specified number of times.
```{r}
rep(x = 1.5, times = 4)
```
#### Random Vector
The `sample()` function takes a random sample from a set of elements. `replace = FALSE` means each element can only be chosen once.
```{r}
sample(1:10, 5, replace = FALSE)
```
With `replace = TRUE`, elements can be chosen multiple times.
```{r}
sample(1:10, 5, replace = TRUE)
```
`runif()` generates random numbers from a uniform distribution.
```{r}
runif(1, min = 0, max = 1)
```
`rnorm()` generates random numbers from a normal distribution.
```{r}
sn1 <- rnorm(4, mean = 0, sd = 1) # Standard normal distribution
sn1
```
#### Unique Vector
The `unique()` function removes duplicate elements from a vector.
```{r}
v1 = c(1, 1, 2, 2, 5, 6)
v1
```
```{r}
unique(v1)
```
### Type Coercion
If you try to mix types, R will **coerce** the elements to the most flexible type in a specific hierarchy: `logical` -> `integer` -> `numeric` -> `character`. For example, mixing numbers and characters will result in a character vector because character is the most flexible type.
```{r}
mixed_vector <- c(1, "apple", 3.5, TRUE)
class(mixed_vector) # All elements are now characters
mixed_vector
```
### Vector Operations
Mathematical and logical operations on vectors are performed element-wise. This is a powerful feature called vectorization, which is much faster than writing a loop.
```{r}
a <- c(1, 2, 3)
b <- c(10, 20, 30)
a + b # Element-wise addition
a * 10 # Scalar multiplication is applied to each element
```
### Subsetting Vectors
You can select elements from a vector using their index inside square brackets `[]`. R uses 1-based indexing.
```{r}
character_vector[2] # Select the second element
numeric_vector[c(1, 3)] # Select the first and third elements
numeric_vector[numeric_vector > 10] # Select elements based on a logical condition
# You can also exclude elements using negative indices
numeric_vector[-2] # Exclude the second element
```
### Append Vector
You can combine vectors by using the `c()` function.
```{r}
x = c(1, 2, 3)
y = c(4, 5, 6)
z = c(x, y)
z
```
### Remove Elements from a Vector
Negative indexing removes elements at the specified positions.
```{r}
x = c(1, 2, 3, 4, 5)
x
```
Remove the first element:
```{r}
x[-1]
```
Remove the last element:
```{r}
x[-length(x)]
```
Remove elements based on a vector of indices:
```{r}
remove = c(2, 4)
x[-remove]
```
### Sort Vector
`sort()` arranges vector elements in ascending or descending order.
```{r}
a = c(2, 4, 6, 1, 4)
sort(a)
```
```{r}
sort(a, decreasing = TRUE)
```
### Vector Length
`length()` returns the number of elements in a vector.
```{r}
length(a)
```
### Vector Calculation
Mathematical functions can be applied to entire vectors.
```{r}
x = c(1, 2, 3, 4, 5)
sum(x)
```
### Select Vector Elements
```{r}
x = c(1, 2, 3, 6, 9, 10)
```
Select the first element:
```{r}
x[1]
```
Select the last element:
```{r}
x[length(x)]
```
Select a range of elements:
```{r}
x[1:3]
```
### Compare Two Vectors
`setdiff(x, y)` finds elements that are in vector `x` but not in vector `y`.
```{r}
xx = c(1, 2, 3, 4)
yy = c(2, 4)
setdiff(xx, yy)
```
### Converting Between Vector Types
`as.*` functions are used to coerce vectors from one type to another.
```{r}
x <- c("a", "g", "b")
y = as.factor(x)
y
```
```{r}
x <- c('123', '44', '222')
y = as.numeric(x)
y
```
# 2. Data Frames
A data frame is a two-dimensional, **heterogeneous** data structure, similar to a spreadsheet or a SQL table. It is the most important data structure for data analysis in R.
- Each column is a vector.
- Each column can have a different data type.
- All columns must have the same length.
## Creating Data Frames
Use the `data.frame()` function or, preferably, the `tibble()` function from the `tidyverse`. Tibbles are a modern take on data frames that are more user-friendly: they don't change variable names or types, and they have a much nicer print method.
```{r}
my_df <- tibble(
id = 1:4,
name = c("Alice", "Bob", "Charlie", "David"),
score = c(95, 82, 78, 91),
is_student = c(TRUE, FALSE, TRUE, FALSE)
)
my_df
```
## Subsetting Data Frames
You can subset data frames in several ways:
- `$` or `[[ ]]` to select a single column by name. This returns the column as a vector.
- `[row, column]` to select specific rows and columns. The result is another data frame (or a vector if you select a single column).
```{r}
# Select the 'name' column (returns a vector)
my_df$name
# Select the first two rows and the 'name' and 'score' columns (returns a tibble)
my_df[1:2, c("name", "score")]
```
For more complex subsetting, it is highly recommended to use the `dplyr` verbs `filter()` and `select()` (covered in a separate guide).
## Data Frame to Matrix
Converting a data frame to a matrix will coerce all elements to the most flexible data type (usually character).
```{r}
mat <- as.matrix(my_df)
mat
```
## Data Frame to Vector
You can extract a single column as a vector using `$` or `[[ ]]` notation.
```{r}
vec = my_df[['name']]
vec
```
# 3. Matrices
A matrix is a two-dimensional, **homogeneous** data structure. All elements must be of the same type. It is most useful for mathematical and statistical operations, such as linear algebra.
### Creating Matrices
Use the `matrix()` function.
```{r}
my_matrix <- matrix(
data = 1:12, # The data to fill the matrix
nrow = 3, # The number of rows
ncol = 4, # The number of columns
byrow = TRUE # Fill the matrix row by row (default is by column)
)
my_matrix
```
### Subsetting Matrices
Subsetting is similar to data frames, using `[row, column]` notation.
```{r}
my_matrix[2, 3] # Element in the 2nd row, 3rd column
my_matrix[1, ] # The entire 1st row
my_matrix[, 4] # The entire 4th column
```
### Matrix Operations
Matrices support matrix algebra operations, such as transposition (`t()`) and matrix multiplication (`%*%`).
```{r}
t(my_matrix) # Transpose the matrix
```
# 4. Lists
A list is a one-dimensional, **heterogeneous** data structure. Unlike vectors, lists can contain elements of different types and even different structures, including other lists, vectors, or data frames. This makes them very flexible, like a container for other objects.
## Creating Lists
Use the `list()` function. It's good practice to name the list elements.
```{r}
# A list is often used to return multiple results from a function
my_model_output <- list(
name = "Linear Model",
coefficients = c(intercept = 2.5, slope = 0.8),
r_squared = 0.85,
data = head(mtcars)
)
my_model_output
```
## Subsetting Lists
Subsetting lists requires understanding the difference between `[` and `[[`:
- `[[index]]` or `[[name]]` extracts a **single element** from the list. The result will have the type of that element.
- `[index]` or `[name]` extracts a **sub-list**. The result is always another list, even if you only select one element.
```{r}
# Extract the 'coefficients' vector (result is a numeric vector)
my_model_output[["coefficients"]]
# Extract a sub-list containing only the 'coefficients' element (result is a list)
my_model_output["coefficients"]
# Use the $ operator as a convenient shortcut for [[name]]
my_model_output$r_squared
```
# 5. Arrays
An array is a multi-dimensional, **homogeneous** data structure. It can have two or more dimensions. A 2D array is essentially a matrix. Arrays are useful for storing data that has more than two dimensions, such as image data (height, width, color channels) or time series data for multiple locations.
## Creating Arrays
Use the `array()` function, specifying the data and the dimensions.
```{r}
# Create a 3D array (2 rows, 3 columns, 2 layers)
my_array <- array(
data = 1:12,
dim = c(2, 3, 2)
)
my_array
```
## Subsetting Arrays
Elements are accessed using `[row, column, dimension]` notation.
```{r}
# Element in the 1st row, 2nd column of the 2nd layer
my_array[1, 2, 2]
# The entire first matrix (1st layer)
my_array[, , 1]
```
# 6. Inspecting Data Structures
Understanding the structure of your data is a critical first step in any analysis. R provides several useful functions for this.
- `str()`: The most useful function. Provides a compact, human-readable summary of any R object.
- `class()`: Returns the high-level class of an object.
- `typeof()`: Returns the internal storage type.
- `length()`: Returns the number of elements in a vector or list.
- `dim()`: Returns the dimensions (e.g., rows and columns) of a data frame, matrix, or array.
- `names()` or `colnames()`: Returns the names of elements or columns.
```{r}
str(my_df)
```
```{r}
str(my_model_output)
```
# 7. Choosing the Right Data Structure
| Use a... | When you have... | Example Use Case |
|----------------|--------------------------------------------------------|-----------------------------------------------------|
| **Vector** | A 1D sequence of the **same data type**. | A single column of data, like ages or names. |
| **Data Frame** | A 2D table with columns of **different data types**. | Your typical dataset for analysis. |
| **Matrix** | A 2D grid of the **same data type**. | A correlation matrix, an image grayscale matrix. |
| **List** | A collection of **different types/structures**. | To return multiple, varied objects from a function. |
| **Array** | A multi-dimensional grid of the **same data type**. | 3D medical imaging data, climate data over time. |
# 8. Reference
- [GeeksforGeeks: Data Structures in R](https://www.geeksforgeeks.org/data-structures-in-r-programming/)
- [Advanced R by Hadley Wickham](https://adv-r.hadley.nz/vectors-chap.html)